《科学》:诺奖得主解读 CRISPR 的十年
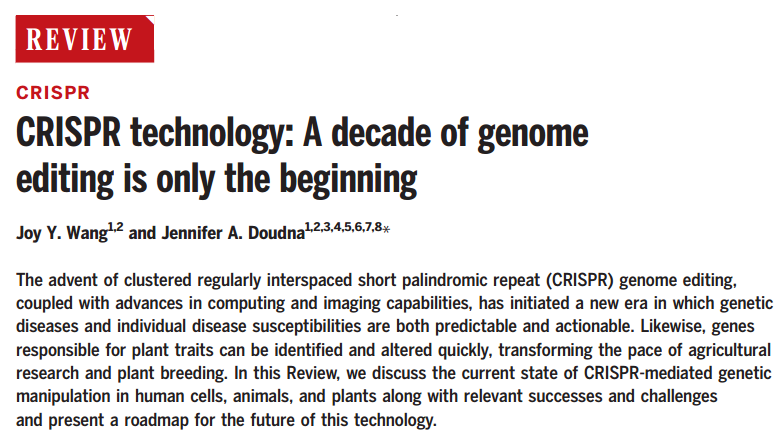
图1 CRISPR 的十年
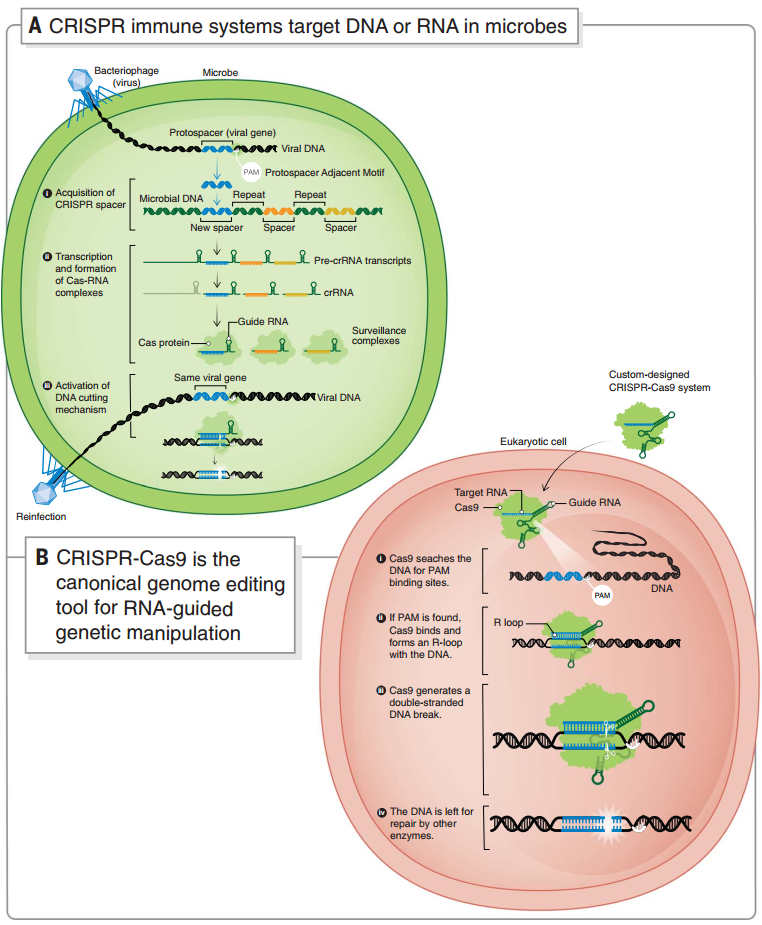
图2 微生物中的 CRISPR 自我防御系统和 CRISPR-Cas9 基因编辑系统
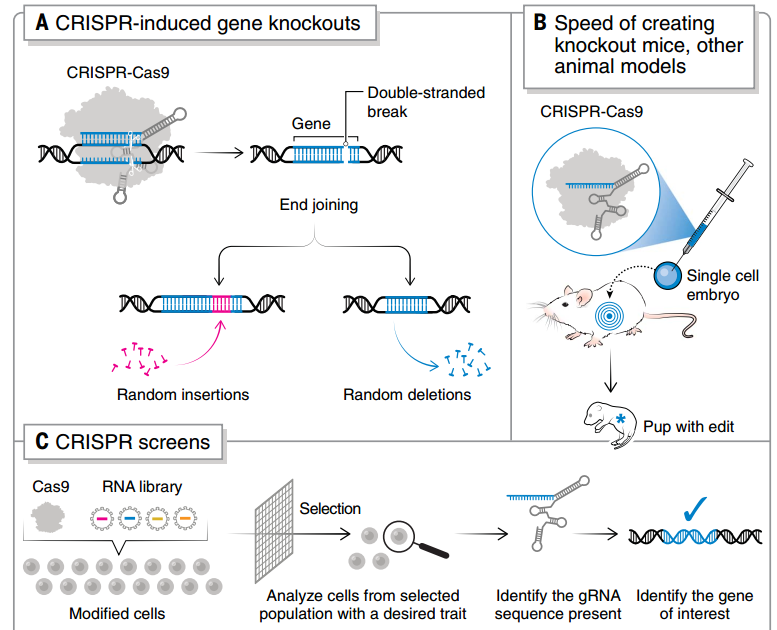
图3 CRISPR 敲除和 CRISPR 筛选的流程
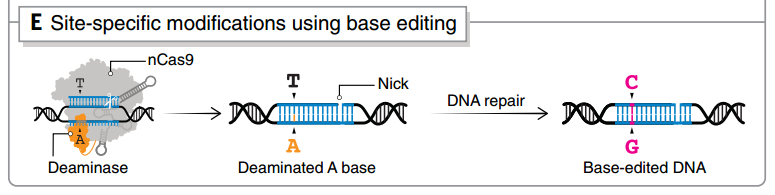
图4 单碱基编辑技术之一base-editing的原理
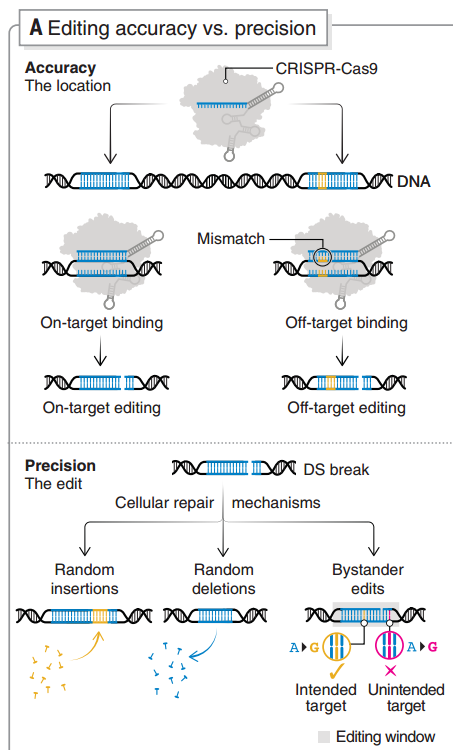
图5 基因编辑特异性和准确性
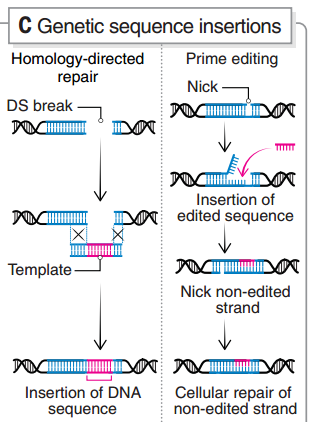
图6 引导编辑的原理
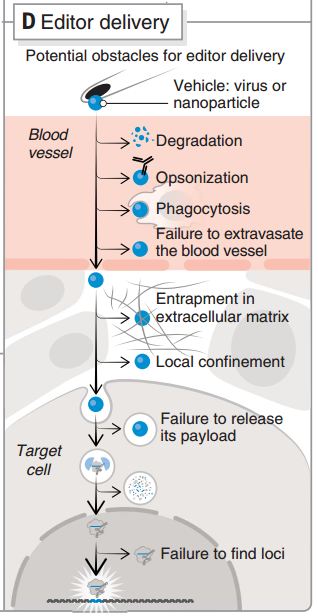
图7 CRISPR 递送过程
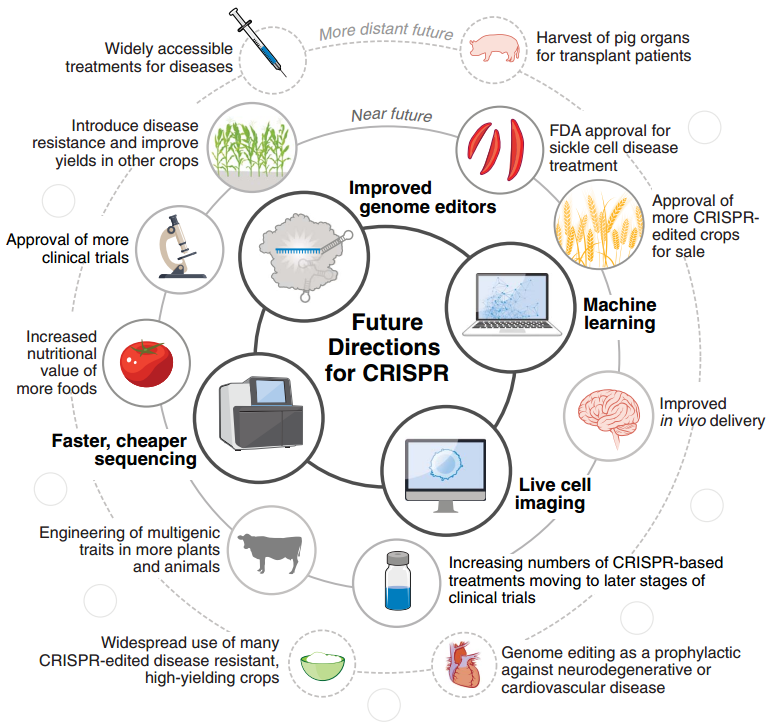
图8 未来的 CRISPR
[2] Y. Ishino, H. Shinagawa, K. Makino, M. Amemura, A. Nakata,Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169,5429–5433 (1987). doi: 10.1128/jb.169.12.5429-5433.1987;pmid: 3316184
[3] F. J. M. Mojica, C. Díez-Villaseñor, J. García-Martínez, E. Soria,Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol.60, 174–182 (2005). doi: 10.1007/s00239-004-0046-3;pmid: 15791728
[4] S. J. J. Brouns et al., Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).doi: 10.1126/science.1159689; pmid: 18703739
[5] L. S. Qi et al., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression.Cell 152, 1173–1183 (2013). doi: 10.1016/j.cell.2013.02.022;pmid: 23452860
[6] J. Y. Wang, P. Pausch, J. A. Doudna, Structural biology of CRISPR-Cas immunity and genome editing enzymes.Nat. Rev. Microbiol. 20, 641–656 (2022). doi: 10.1038/s41579-022-00739-4; pmid: 35562427
[7] H. Yang, H. Wang, R. Jaenisch, Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9, 1956–1968 (2014). doi: 10.1038/nprot.2014.134; pmid: 25058643
[8] H. Mou, Z. Kennedy, D. G. Anderson, H. Yin, W. Xue, Precision cancer mouse models through genome editing with CRISPRCas9. Genome Med. 7, 53 (2015). doi: 10.1186/s13073-015-0178-7; pmid: 26060510
[9] H. Lee, D. E. Yoon, K. Kim, Genome editing methods in animal models. Anim. Cells Syst. 24, 8–16 (2020). doi: 10.1080/19768354.2020.1726462; pmid: 32158611
[10] A. Katti, B. J. Diaz, C. M. Caragine, N. E. Sanjana, L. E. Dow,CRISPR in cancer biology and therapy. Nat. Rev. Cancer 22,259–279 (2022). doi: 10.1038/s41568-022-00441-w;pmid: 35194172
[11] N. E. Sanjana, O. Shalem, F. Zhang, Improved vectors and genome-wide libraries for CRISPR screening. Nat.Methods 11, 783–784 (2014). doi: 10.1038/nmeth.3047;pmid: 25075903
[12] L. Przybyla, L. A. Gilbert, A new era in functional genomics screens. Nat. Rev. Genet. 23, 89–103 (2022). doi: 10.1038/s41576-021-00409-w; pmid: 34545248
[13] G. M. Findlay et al., Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018).doi: 10.1038/s41586-018-0461-z; pmid: 30209399
[14] L. W. Koblan et al., In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature 589, 608–614 (2021). doi: 10.1038/s41586-020-03086-7;pmid: 33408413
[15] K. Kingwell, Base editors hit the clinic. Nat. Rev. Drug Discov.21, 545–547 (2022). doi: 10.1038/d41573-022-00124-z;pmid: 35831515
[16] W. Chen et al., Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated doublestrand break repair. Nucleic Acids Res. 47, 7989–8003(2019). doi: 10.1093/nar/gkz487; pmid: 31165867
[17] C. D. Richardson, G. J. Ray, M. A. DeWitt, G. L. Curie,J. E. Corn, Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344(2016). doi: 10.1038/nbt.3481; pmid: 26789497
[18] A. Lomova et al., Improving gene editing outcomes in human hematopoietic stem and progenitor cells by temporal control of DNA repair. Stem Cells 37, 284–294 (2019).doi: 10.1002/stem.2935; pmid: 30372555
[19] X. Ling et al., Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates.Sci. Adv. 6, eaaz0051 (2020). doi: 10.1126/sciadv.aaz0051;pmid: 32494588
[20] M. Arbab et al., Determinants of base editing outcomes from target library analysis and machine learning. Cell 182, 463–480.e30 (2020). doi: 10.1016/j.cell.2020.05.037; pmid: 32533916
[21]M. M. Hassan, G. Yuan, J.-G. Chen, G. A. Tuskan, X. Yang,Prime editing technology and its prospects for future applications in plant biology research. BioDesign Research 2020, 1–14 (2020). doi: 10.34133/2020/9350905
[22] J. D. Gillmore et al., CRISPR-Cas9 In vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502(2021). doi: 10.1056/NEJMoa2107454; pmid: 34215024
[23] F. D. Urnov, The Cas9 hammer-and sickle: A challenge for genome editors. CRISPR J. 4, 6–13 (2021). doi: 10.1089/crispr.2021.29120.fur; pmid: 33616446
[24] K. Musunuru et al., In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593,429–434 (2021). doi: 10.1038/s41586-021-03534-y;pmid: 34012082
[25] S. Li et al., Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460(2022). doi: 10.1038/s41586-022-04395-9;pmid: 35140403
 |
 |
 |
| 官网:www.cxbio.com | 微信服务号:iseebio | 微博:seebiobiotech |
 |
 |
 |
| 商城:mall.seebio.cn | 微信订阅号:seebiotech | 泉养堂:www.canmedo.com |
相关资讯
- MIT神经学大牛关于阿尔茨海默病的超级颠覆研究
- 杭师大:miRNA调控水稻叶片衰老
- CTI:深度揭示COVID-19患者的免疫反应特征
- 8分钟检测A群链球菌感染 Alere分子诊断试剂获FDA批准
- 皮肤素硫酸盐低聚糖标准品
- 「亚星官网yaxin111」专业提供美国NU-CHEK【脂肪酸标准品】
- 孩子最喜欢吃的彩虹糖可致DNA改变!伤害大脑、肝脏、肾脏……
- 环氧化磁珠
- 揭开白藜芦醇的延年益寿之谜
- 干细胞“种”出血管化腐生肌 中山医院保肢率达91.2%
新进产品
同类文章排行
- 《科学》:诺奖得主解读 CRISPR 的十年
- 《Nature》甲流当季,大脑如何感知感染以及“布洛芬”没效果的原因?
- 修改mRNA或可治疗阿尔茨海默病
- Immunity:免疫系统的“马拉松选手”
- 《PNAS》科学家解开细胞存活之谜
- 大脑越用越“废”?Nature发现神经元DNA修复机制,或推动相关疾病研究进展
- 干细胞疗法创新:新的水凝胶维持干细胞存活 可修复小鼠受伤脑组织
- 肺细胞如何保护自己免受RNA病毒感染?
- 《柳叶刀》:首次成功利用CAR-T细胞治疗严重肌肉炎症
- Science子刊:对过敏性哮喘的保护—先天淋巴样细胞
资讯文章
您的浏览历史









